Tree building for 153 Coronavirus genomes¶
- Notebook version:
v0.0.1 - Created by: Dr. Hiren Ghosh,
Imperial BRC Genomics Facility - Maintained by:
Imperial BRC Genomics Facility - Docker image: imperialgenomicsfacility/viral-genome-analysis-notebooks
- Github repository: imperial-genomics-facility/viral-genome-notebook-image
- Created on:
2020-April-21 14:42 - Contact us: Imperial BRC Genomics Facility
- License: Apache License 2.0
Fetch data¶
Download the aligned corona virus genomes from Genexa
[1]:
## Downloading aligned fasta format
!wget https://storage.googleapis.com/sars-cov-2/MSA_SARS2_20200329.fasta
--2020-04-02 18:49:21-- https://storage.googleapis.com/sars-cov-2/MSA_SARS2_20200329.fasta
Resolving storage.googleapis.com (storage.googleapis.com)... 74.125.129.128, 2607:f8b0:4001:c03::80
Connecting to storage.googleapis.com (storage.googleapis.com)|74.125.129.128|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 4668949 (4.5M) [application/octet-stream]
Saving to: 'MSA_SARS2_20200329.fasta'
MSA_SARS2_20200329. 100%[===================>] 4.45M --.-KB/s in 0.06s
2020-04-02 18:49:21 (70.8 MB/s) - 'MSA_SARS2_20200329.fasta' saved [4668949/4668949]
[37]:
## DOwnloading data in Clustal format
!wget -O MSA_SARS2_20200329.aln https://storage.googleapis.com/sars-cov-2/MSA_SARS2_20200329.clw
--2020-04-02 19:10:59-- https://storage.googleapis.com/sars-cov-2/MSA_SARS2_20200329.clw
Resolving storage.googleapis.com (storage.googleapis.com)... 172.217.214.128, 2607:f8b0:4001:c07::80
Connecting to storage.googleapis.com (storage.googleapis.com)|172.217.214.128|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 5932931 (5.7M) [application/octet-stream]
Saving to: 'MSA_SARS2_20200329.aln'
MSA_SARS2_20200329. 100%[===================>] 5.66M --.-KB/s in 0.1s
2020-04-02 19:10:59 (57.8 MB/s) - 'MSA_SARS2_20200329.aln' saved [5932931/5932931]
Build phylogenetic tree with FastTree¶
[2]:
!FastTree -nt -gtr < MSA_SARS2_20200329.fasta >MSA_SARS2_20200329.tree
FastTree Version 2.1.10 Double precision (No SSE3)
Alignment: standard input
Nucleotide distances: Jukes-Cantor Joins: balanced Support: SH-like 1000
Search: Normal +NNI +SPR (2 rounds range 10) +ML-NNI opt-each=1
TopHits: 1.00*sqrtN close=default refresh=0.80
ML Model: Generalized Time-Reversible, CAT approximation with 20 rate categories
Ignored unknown character D (seen 1 times)
Ignored unknown character H (seen 13 times)
Ignored unknown character K (seen 1 times)
Ignored unknown character R (seen 1 times)
Ignored unknown character S (seen 3 times)
Ignored unknown character V (seen 1 times)
Ignored unknown character W (seen 3 times)
Ignored unknown character X (seen 42 times)
Ignored unknown character Y (seen 17 times)
Initial topology in 2.67 seconds0 of 123 126 seqs (at seed 100)
Refining topology: 28 rounds ME-NNIs, 2 rounds ME-SPRs, 14 rounds ML-NNIs
Total branch-length 0.010 after 19.69 sec, 101 of 124 splits, 0 changes ax delta 0.000)
WARNING! This alignment consists of closely-related and very-long sequences.
WARNING! FastTree (or other standard maximum-likelihood tools)
may not be appropriate for aligments of very closely-related sequences
like this one, as FastTree does not account for recombination or gene conversion
ML-NNI round 1: LogLk = -45578.360 NNIs 57 max delta 6.62 Time 27.53ges (max delta 6.617)
GTR Frequencies: 0.2990 0.1837 0.1962 0.3211ep 12 of 12
GTR rates(ac ag at cg ct gt) 0.4703 1.4001 0.5361 0.6047 2.2160 1.0000
Switched to using 20 rate categories (CAT approximation)20 of 20
Rate categories were divided by 0.626 so that average rate = 1.0
CAT-based log-likelihoods may not be comparable across runs
Use -gamma for approximate but comparable Gamma(20) log-likelihoods
ML-NNI round 2: LogLk = -44402.405 NNIs 34 max delta 0.00 Time 89.79ges (max delta 0.000)
Turning off heuristics for final round of ML NNIs (converged)
ML-NNI round 3: LogLk = -44401.762 NNIs 27 max delta 0.55 Time 104.20 (final)delta 0.546)
Optimize all lengths: LogLk = -44401.740 Time 107.77
Total time: 128.59 seconds Unique: 126/153 Bad splits: 0/123rnal splits
Plot trees¶
[3]:
import os
import requests
import json
import time
from ete3 import Tree, TreeStyle,SeqMotifFace
from PIL import Image
os.environ['QT_QPA_PLATFORM']='offscreen'
[4]:
t = Tree("MSA_SARS2_20200329.tree")
[9]:
## plain tree
t.render("%%inline")
[9]:

[32]:
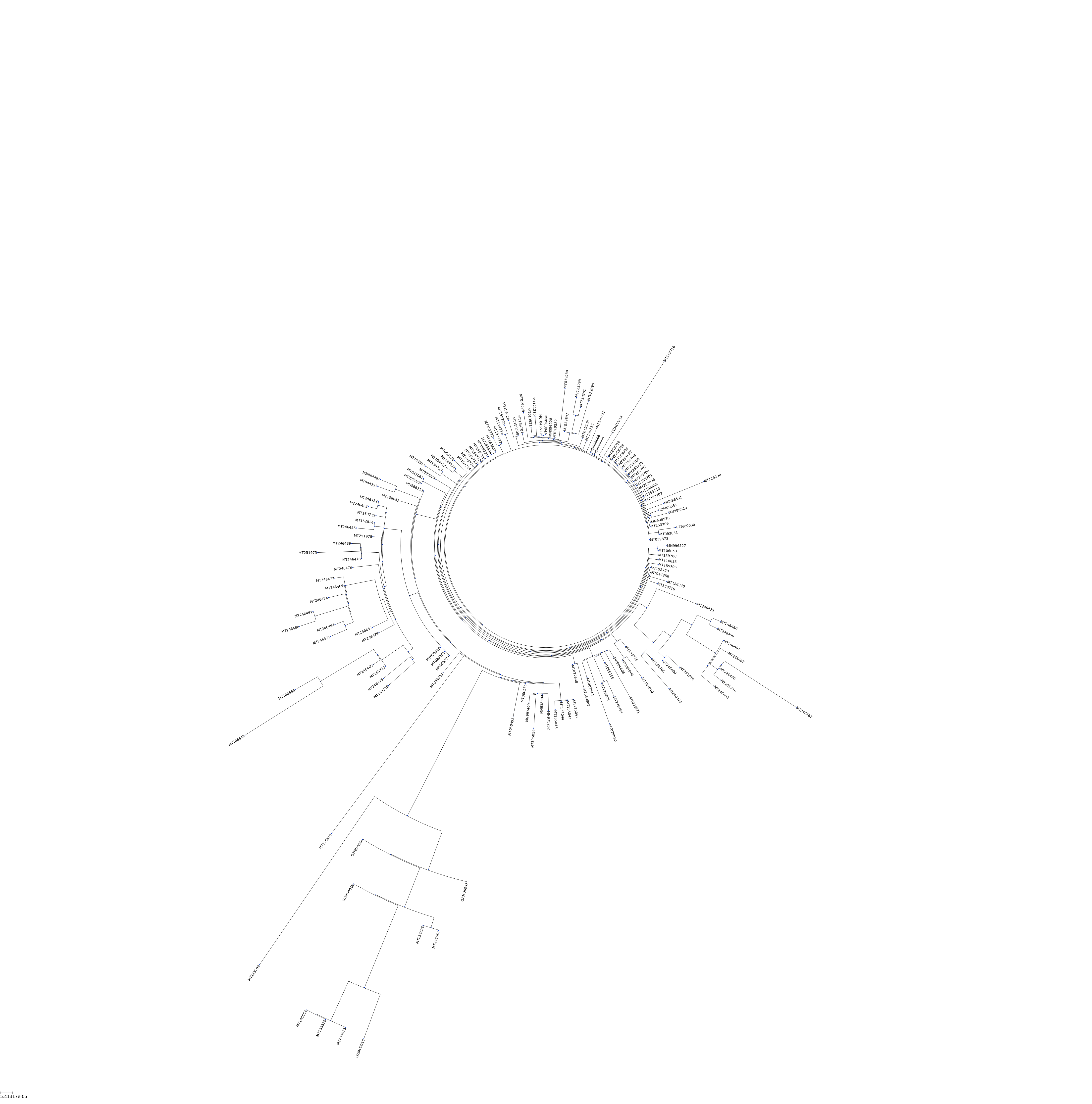
## circular tree
ts = TreeStyle()
ts.mode = "c"
#ts.scale = 20
t.render("%%inline",tree_style=ts)
[32]:
[12]:
## tree with branch length
ts = TreeStyle()
ts.show_leaf_name = True
ts.show_branch_length = True
ts.show_branch_support = True
t.render("%%inline",tree_style=ts)
[12]:
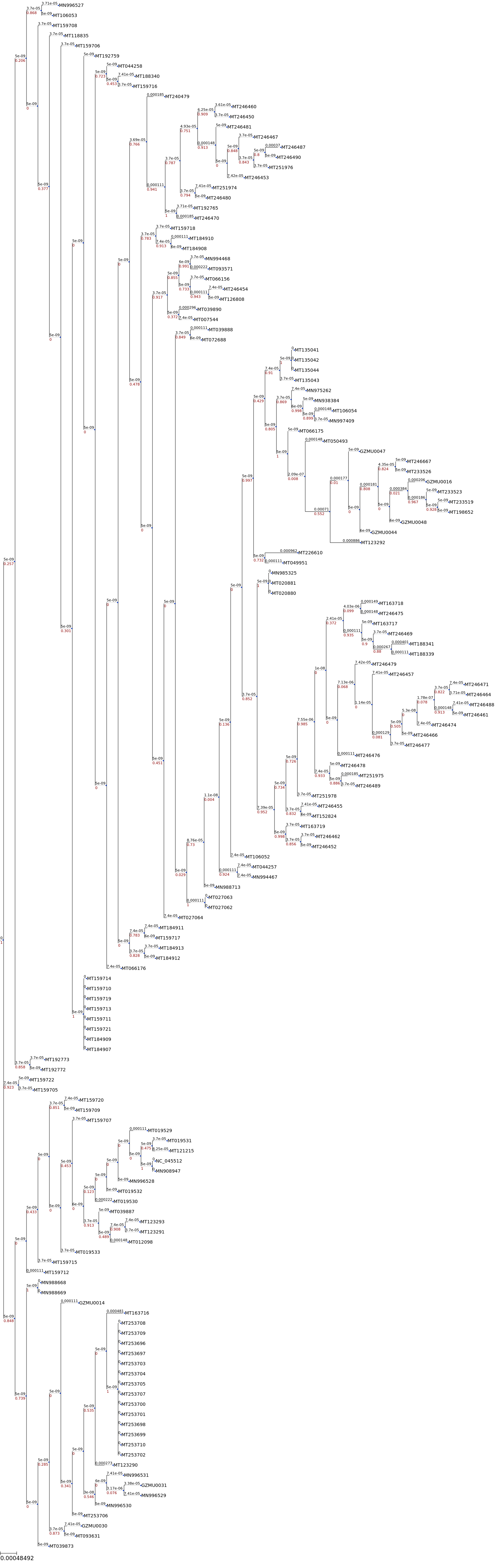
[31]:
## 180 deg circular tree
ts = TreeStyle()
ts.show_leaf_name = True
ts.mode = "c"
ts.arc_start = -180 # 0 degrees = 3 o'clock
ts.arc_span = 180
t.render("180_deg_circular_tree.png",tree_style=ts)
t.render("%%inline",tree_style=ts)
[33]:
## read aligned fastq file
fasta_data = dict()
with open('MSA_SARS2_20200329.fasta','r') as fp:
header = ''
fasta_list = list()
for line in fp:
line = line.strip()
if line.startswith('>'):
if header != '':
fasta_data.update({header:''.join(fasta_list)})
header = line.split()[0].replace('>','')
fasta_list = list()
else:
fasta_list.append(line)
fasta_data.update({header:''.join(fasta_list)})
[35]:
## plot tree with alignment
for seq_id,seq in fasta_data.items():
seqFace = SeqMotifFace(seq, gapcolor="red")
(t & "{0}".format(seq_id)).add_face(seqFace, 0, "aligned")
ts = TreeStyle()
ts.tree_width = 100
t.render("tree_with_aln.png", tree_style=ts);
Reference¶
- Price, M.N., Dehal, P.S., and Arkin, A.P. (2009) FastTree: Computing Large Minimum-Evolution Trees with Profiles instead of a Distance Matrix. Molecular Biology and Evolution 26:1641-1650, doi:10.1093/molbev/msp077.
- Price, M.N., Dehal, P.S., and Arkin, A.P. (2010) FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE, 5(3):e9490. doi:10.1371/journal.pone.0009490.
- ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Jaime Huerta-Cepas, Francois Serra and Peer Bork. Mol Biol Evol 2016; doi: 10.1093/molbev/msw046
[ ]: